Demystifying APIs for Researchers
2024-01-24
Learning Objectives
- Understand what an API (application programming interface) is
- Get a sense of what kinds of data are available via APIs
- Understand how to get data from an API into R programmatically
Slides: https://cct-datascience.quarto.pub/demystifying-apis-slides/
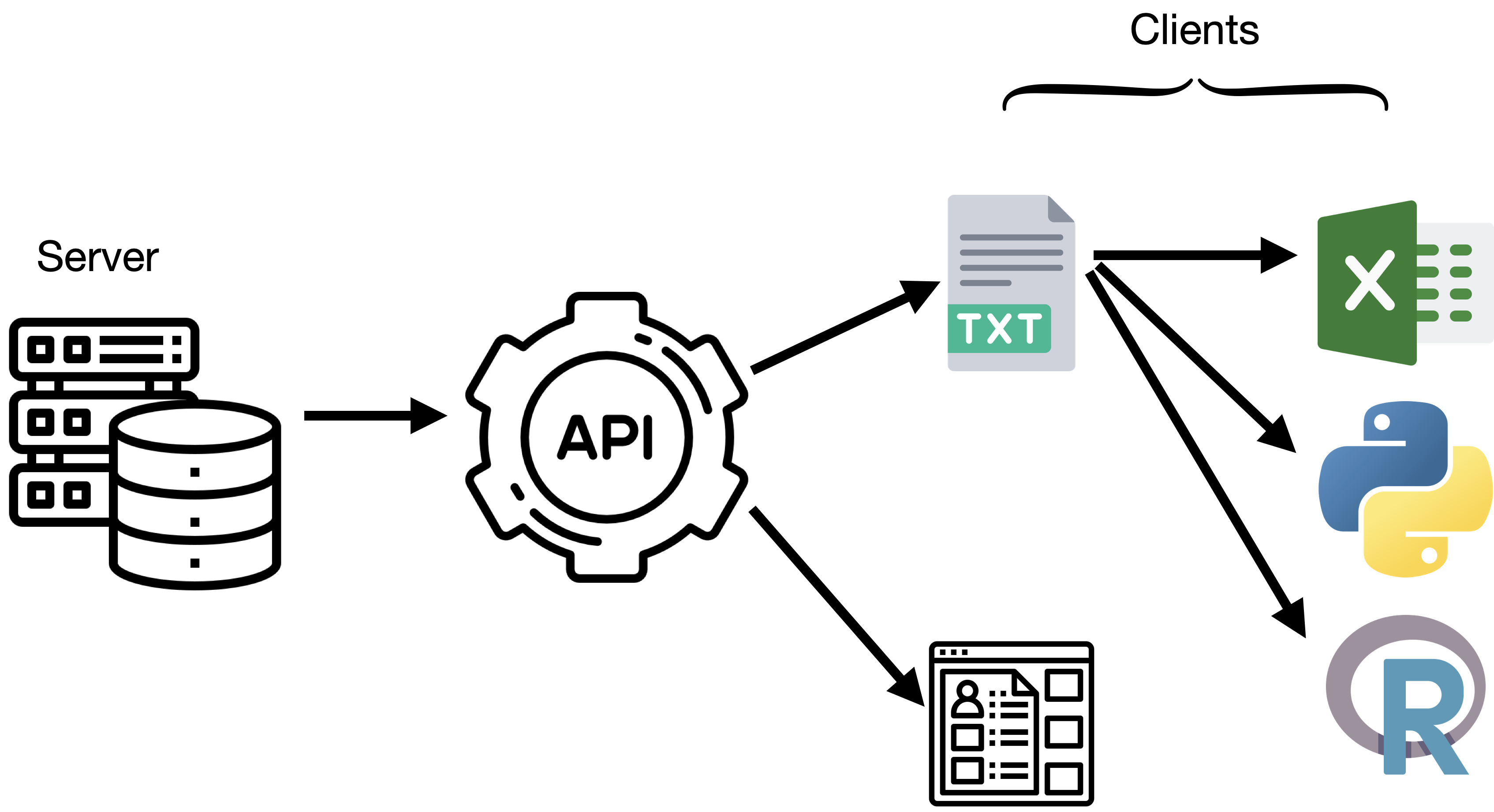
What is an API
Getting data on the web
- Made for humans
- Point and click
E.g. CAS Common Chemistry
- Search for the name of a chemical (e.g. hexanol)
- Click on a result to get more info
What’s missing?
There’s no easy way to get the results in a ready-to-analyze form!
Getting data for analysis
This is where an API is useful
- Made for machines
- Programmable
APIs exist for many data portals
Getting data directly from an API
Every API is a little different, but here the API equivalent to https://commonchemistry.cas.org/results?q=hexanol is https://commonchemistry.cas.org/api/search?q=hexanol.
(I’ll show you how I figured this out in a bit)

What am I looking at?
The result of https://commonchemistry.cas.org/api/search?q =hexanol is in a format called JSON. It’s made of nested key: value pairs. For example
Structure of an API request

Note
This is a fairly standard API, but not all APIs take requests structured in the same way! You’ll have to find the documentation for each particular API to figure out how to build a query.
How to know how to use an API
Let’s look at some API documentation: https://commonchemistry.cas.org/api-overview
What endpoints are available? What queries can you use?
Note
This API only allows one “method”—GET. Other methods exist (e.g. POST), but GET is the most common.
Building an API request in R
Reproducible queries
Iterate over many queries
Create re-usable functions
1. Create a request
Start a request with the domain
2. Perform the request
3. Get the results
List of 2
$ count : int 2
$ results:List of 2
..$ :List of 3
.. ..$ rn : chr "111-27-3"
.. ..$ name : chr "1-Hexanol"
.. ..$ image: chr "<svg width=\"184.65\" viewBox=\"0 0 6155 1002\" text-rendering=\"auto\" stroke-width=\"1\" stroke-opacity=\"1\""| __truncated__
..$ :List of 3
.. ..$ rn : chr "25917-35-5"
.. ..$ name : chr "Hexanol"
.. ..$ image: chr "<svg width=\"142.29\" viewBox=\"0 0 4743 3165\" text-rendering=\"auto\" stroke-width=\"1\" stroke-opacity=\"1\""| __truncated__Live Demo
Other useful functions
req_throttle()for obeying API rate limitsreq_retry()for automatically retrying requests that failreq_oauth_auth_code()and associated helpers for APIs that require access tokens or passwords. See this OAuth article.
Don’t re-invent the wheel!
Many APIs have an R package or Python library to access them. Look for these before writing your own
httr2code!rOpenSci has over 100 data access R packages: https://ropensci.org/packages/data-access/
Getting Help
CCT Data Science drop-in hours: datascience.cct.arizona.edu/drop-in-hours
Email us: cct-datascience@arizona.edu
Book an appointment: datascience.cct.arizona.edu/people
Data Science Incubator program: datascience.cct.arizona.edu/cct-data-science-incubator
Title slide API icon by Lordicon.com
