Crafting Publication Quality Data Visualizations with ggplot2
Learning Objectives
- Customize color palettes with accessibility in mind
- Customize legends
- Customizing axes (titles, labels, breaks)
- Customize appearance of plots with themes
- Arrange multi-panel figures
- Save high resolution or vector formats
Required Packages
Take a moment to check if these load and install them if you need to.
Journal Requirements
Journals often require certain modifications to your plots to make them publication-ready
- High resolution
- Specific file types (TIFF, EPS, PDF are common)
- Figure size limits
- Font size suggestions
Not required, but good practice
Other modifications to the appearance of your plot are a good idea, but less often required by journals or reviewers
- Colorblind accessible colors
- Grey scale friendly colors
- Perceptually-even colors
- Screen-reader compatible
- High data-ink ratio (simplify plot, within reason)
- Arrangement of related plots into multi-panel figures
Example plot 1

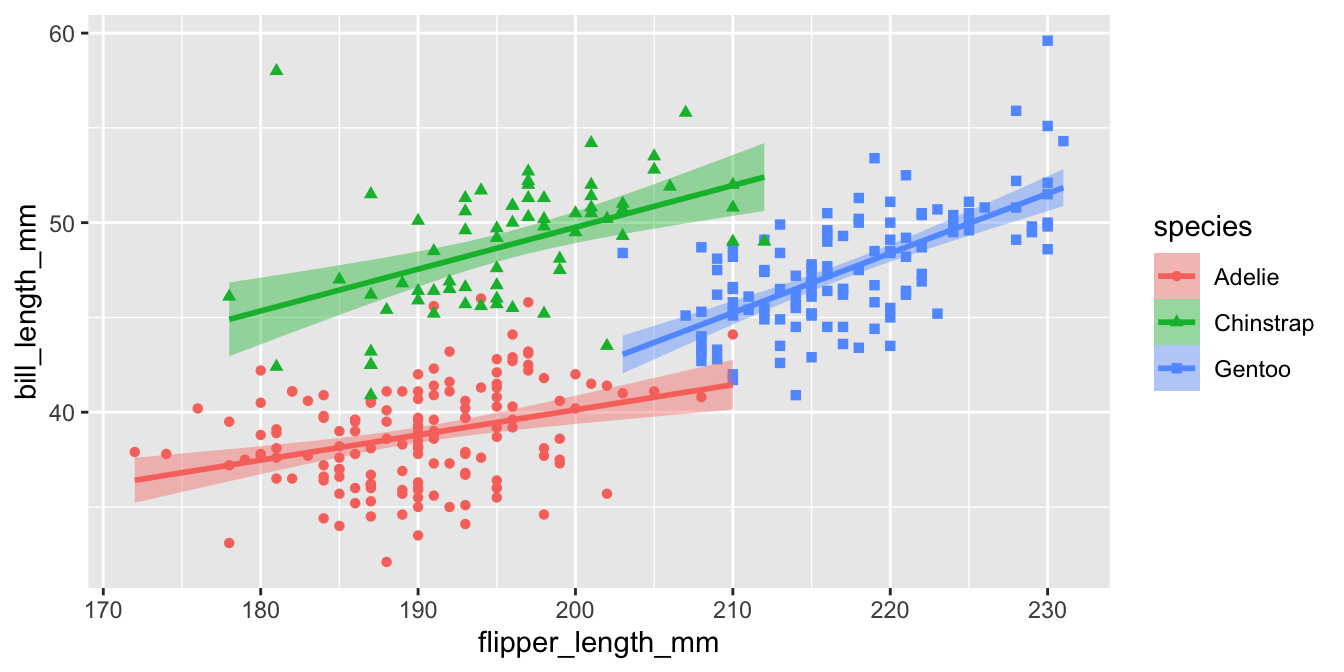
Example plot 2
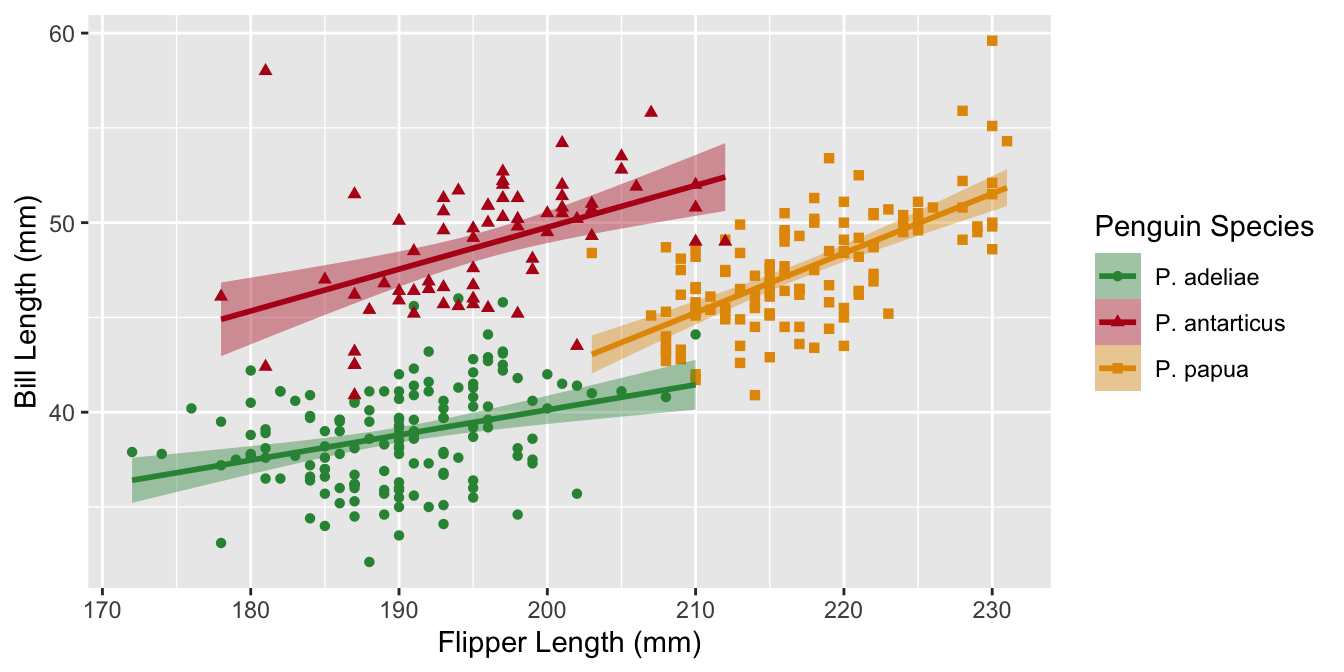
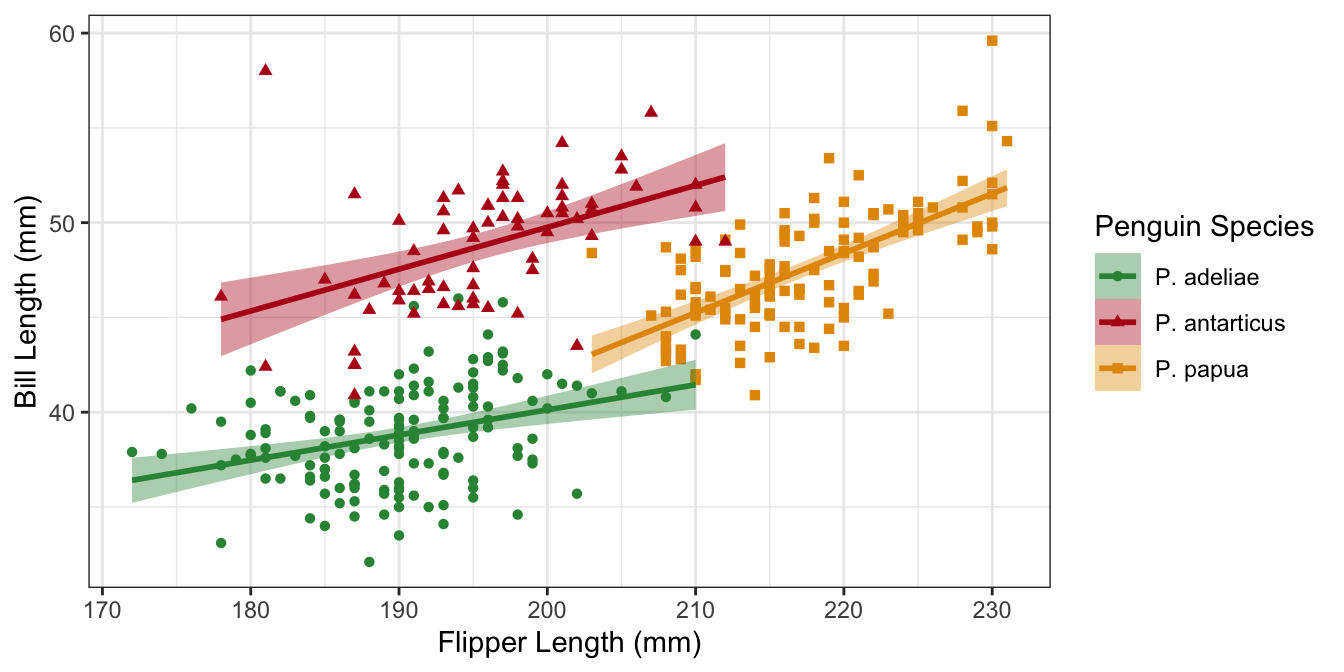
Example plot 3
Custom Colors
Color Palettes
Choose a color palette that is:
- Colorblind friendly
- Greyscale friendly
- Perceptually even
- High contrast (with background & within palette)
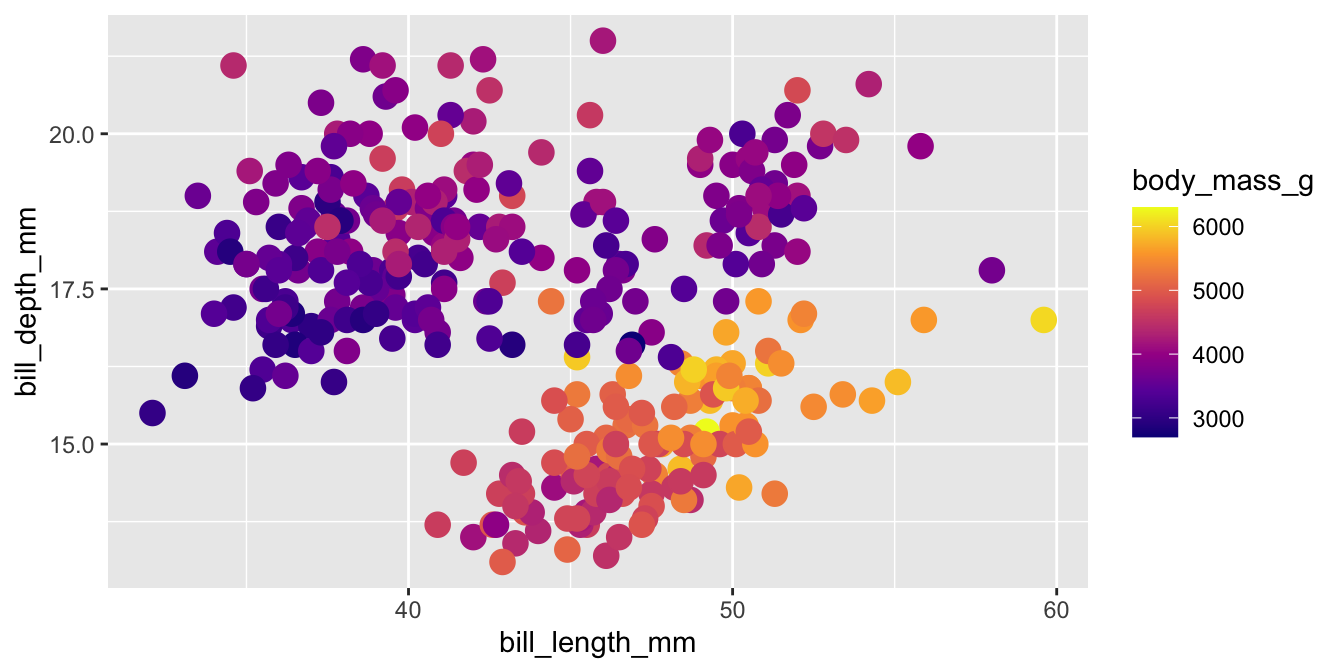
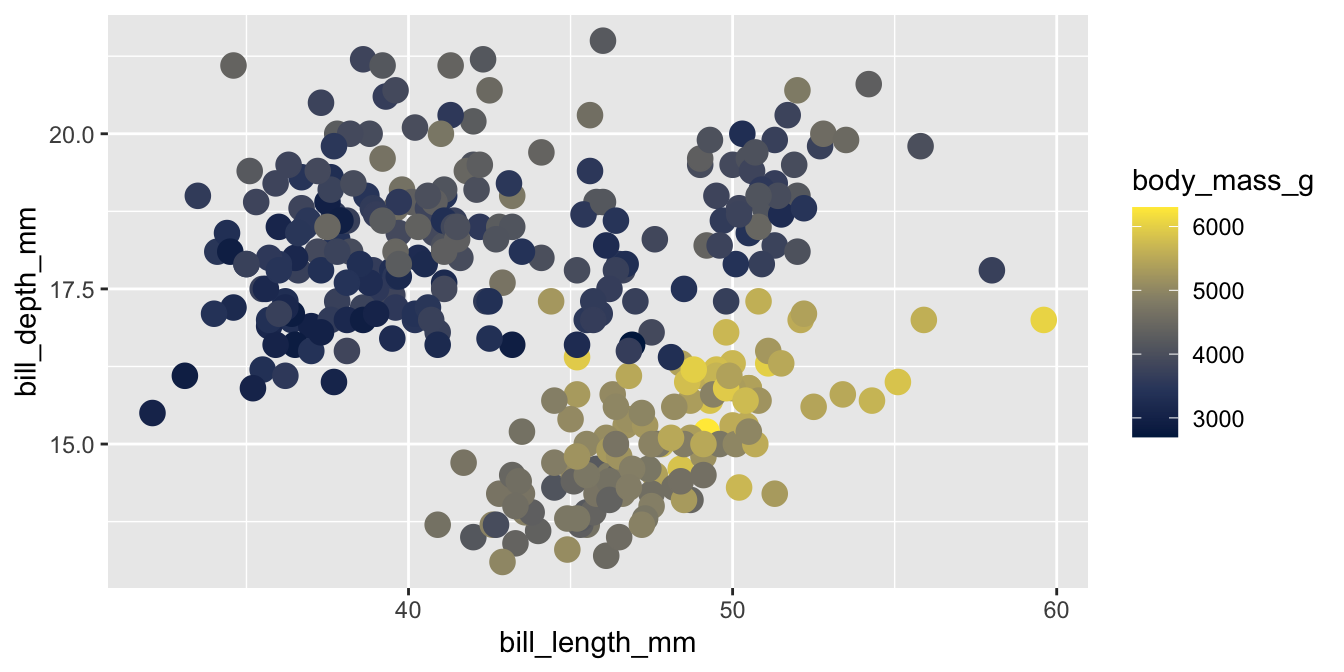
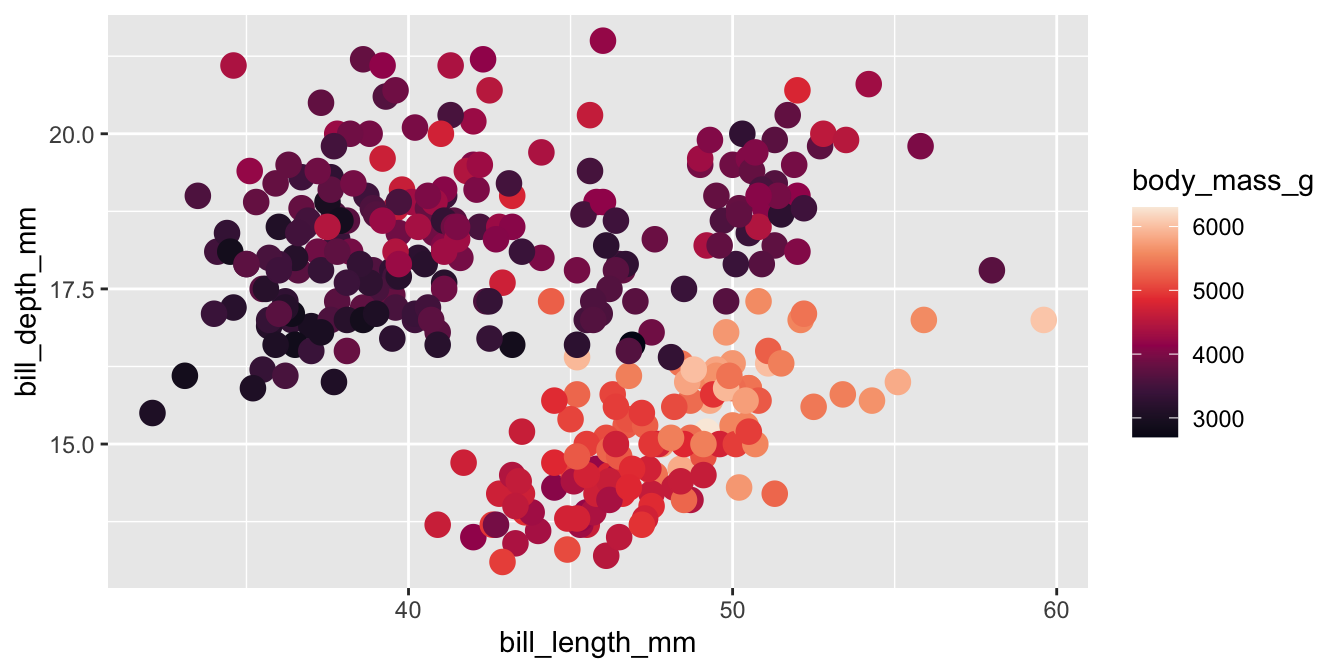
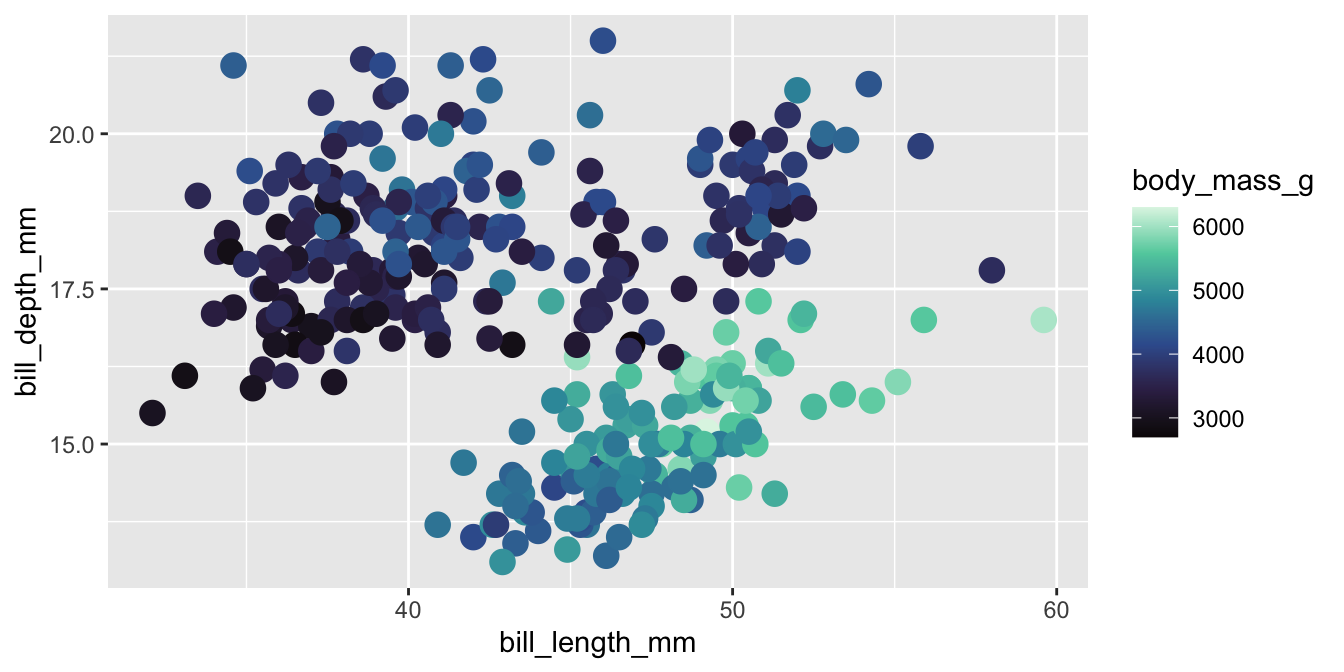
Viridis
The viridis color palettes meet most of these criteria and are built-in to ggplot2. They are available with scale_fill_viridis_*() and scale_color_viridis_*() functions.
Viridis variants
Other viridis palettes are available by changing option in the scale function
Code

Viridis customization
The upper end of viridis palettes tends to be very bright yellow. You can limit the range of colors used with the begin and end arguments
Viridis for discrete data
The viridis palette can be used for discrete / categorical data with scale_color_viridis_d().
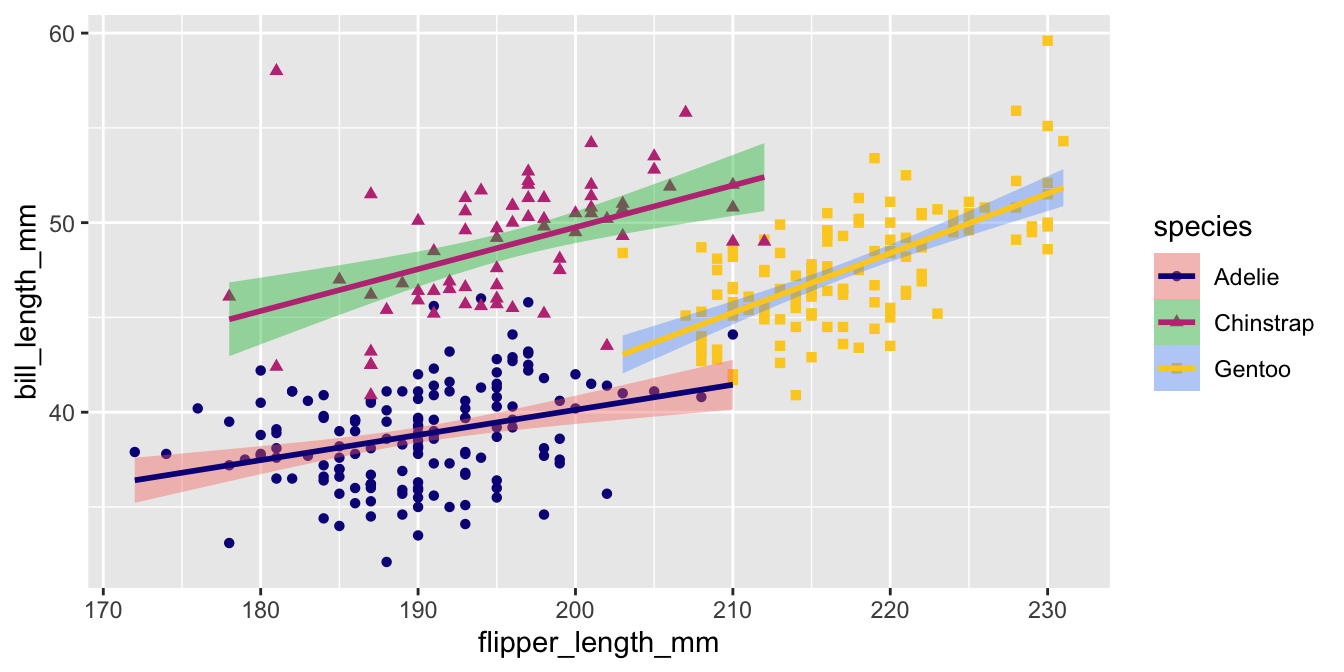
Uh oh!
This only applied the new palette to the color aesthetic!
Applying palettes to multiple aesthetics
Usually color and fill are mapped to the same data. You can add both scale_color_*() and scale_fill_*() to a plot OR you can use the aesthetics argument.
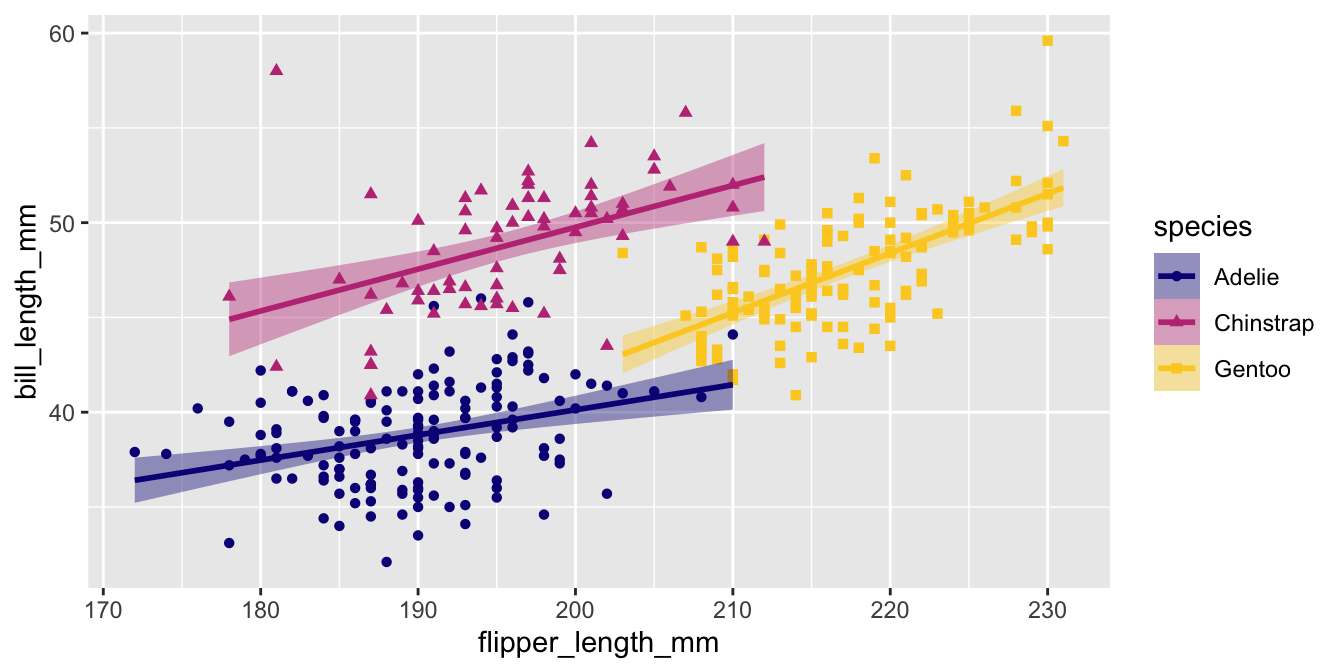
Other color palettes
There are many places to get additional color palettes.
A few of my favorites:
Activity
Let’s find a palette we like using cols4all::4a_gui()
Manual color palettes
You can always use your own colors using scale_color_manual() if you know the hex codes.
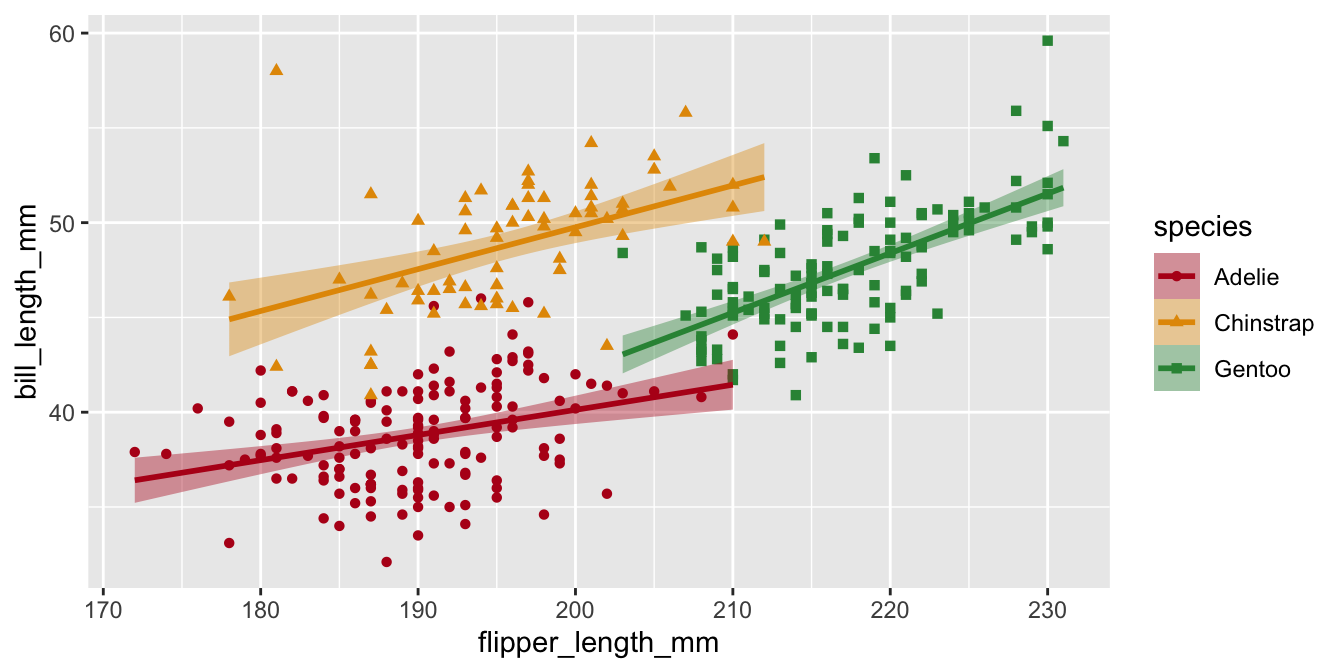
Manual color palettes
Use a named vector to specify which colors go with which factor level
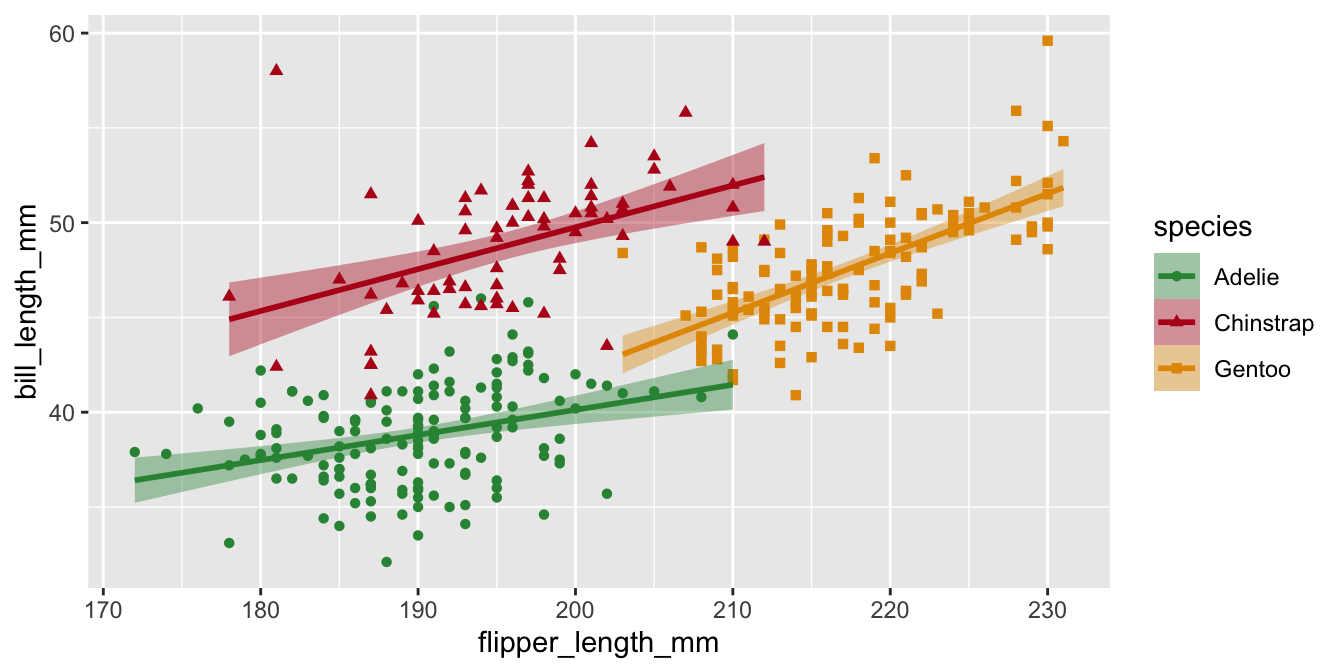

Legends
Legend titles
We can set the name for scales a few ways: with labs() or with the name= argument of the scale.
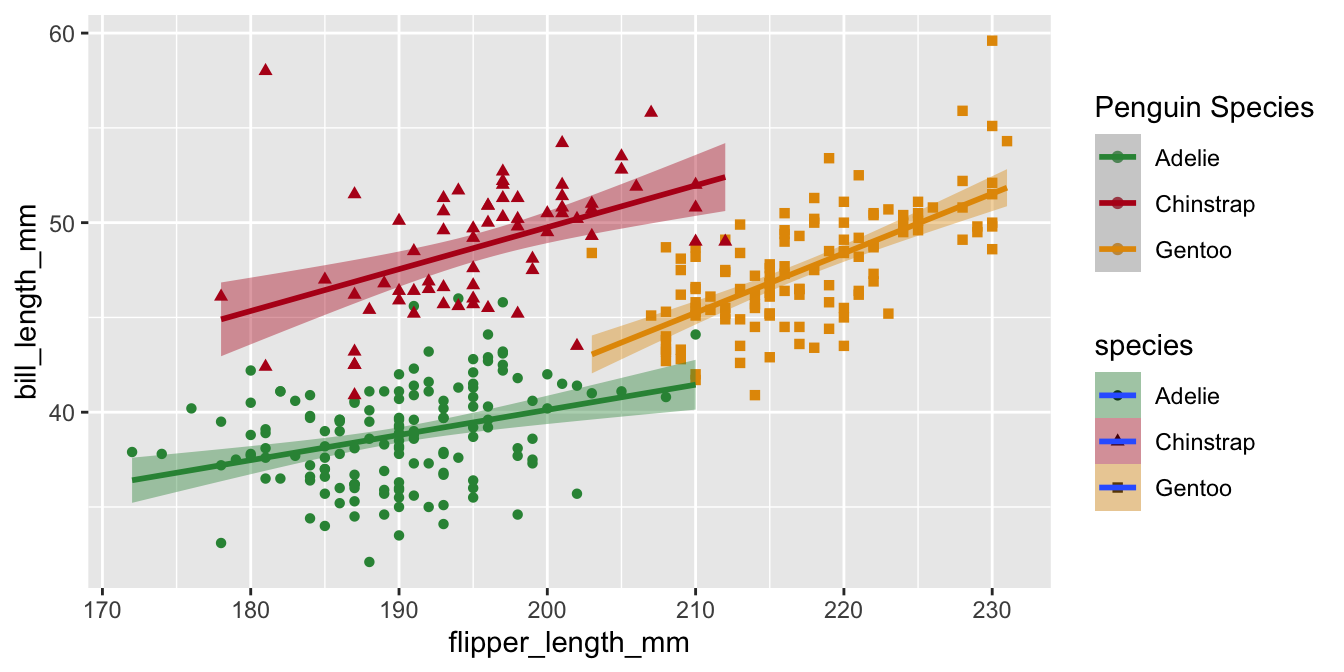
Legend titles
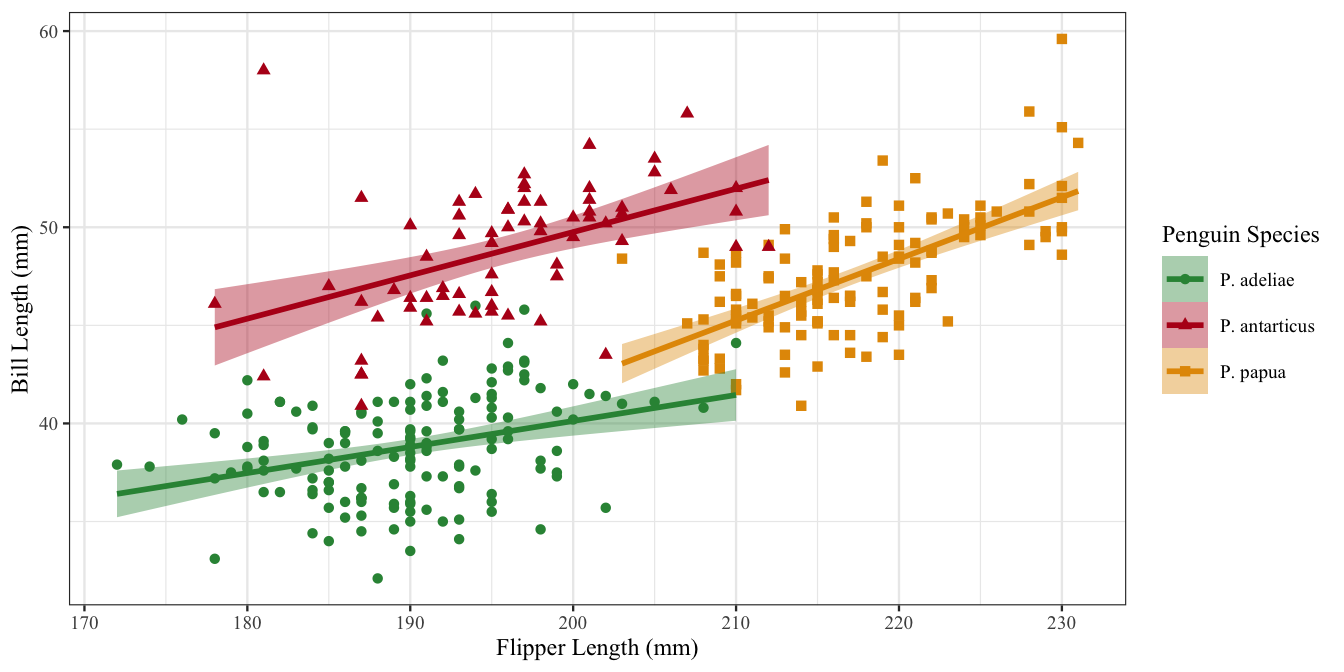
Legends for scales with the same name will be combined if possible
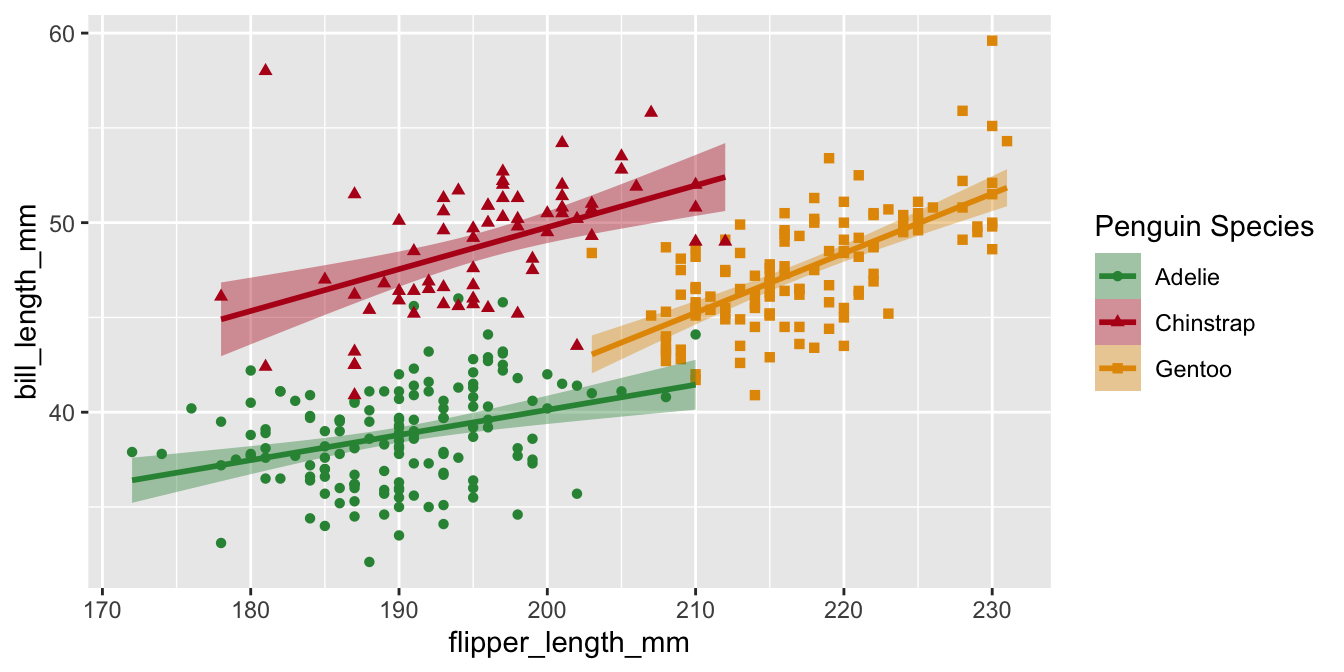
Legend titles
Let’s do the same for p3
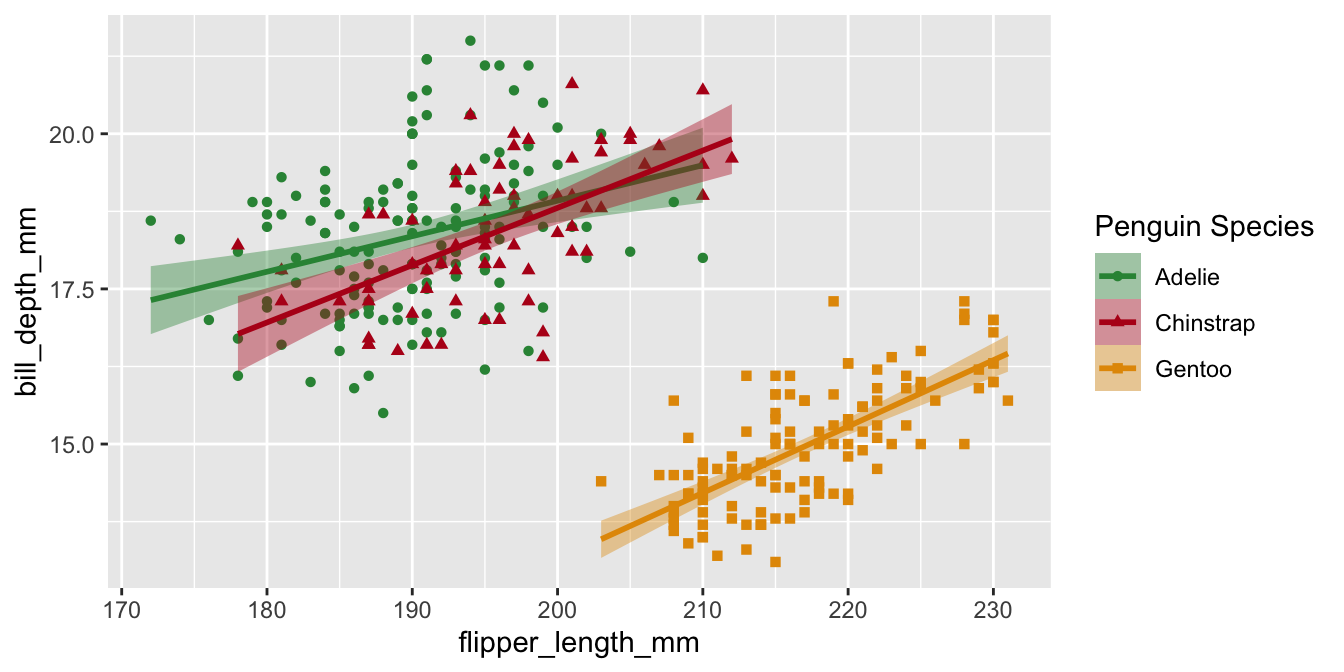
Legend labels
What if we want to use the Latin name for the penguin species? We can use the labels argument and a named vector.
Legend labels
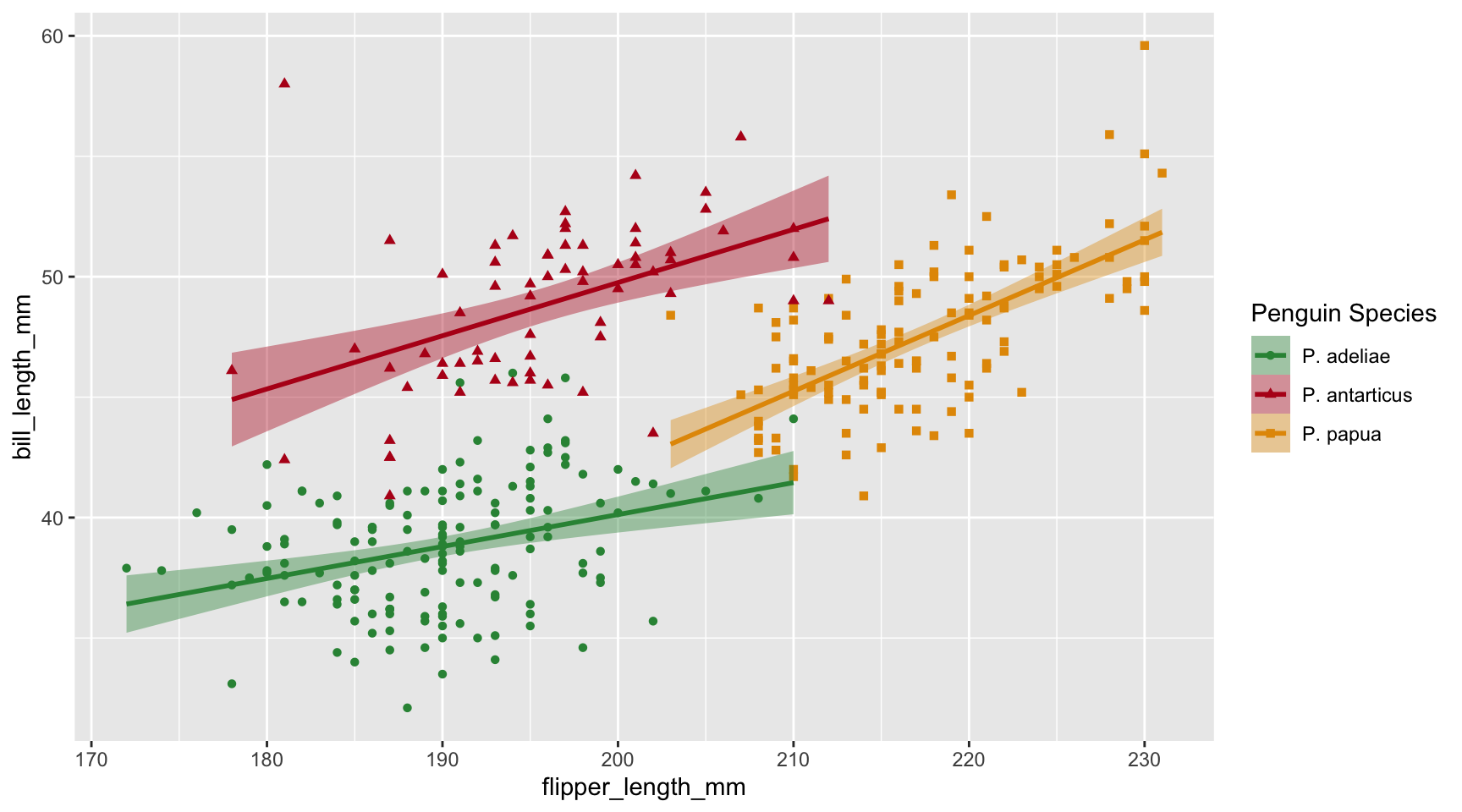
Legend labels
Let’s do the same for p3
Tip
If there are many aesthetics that map to the same variable, it might be easier to change the factor levels in the data once instead of inside of every scale
Legend labels
Applying what we learned
Let’s apply what we learned to p1 to capitalize the words in the legend
Which
scale_function?Which argument changes legend title?
Which argument changes labels?
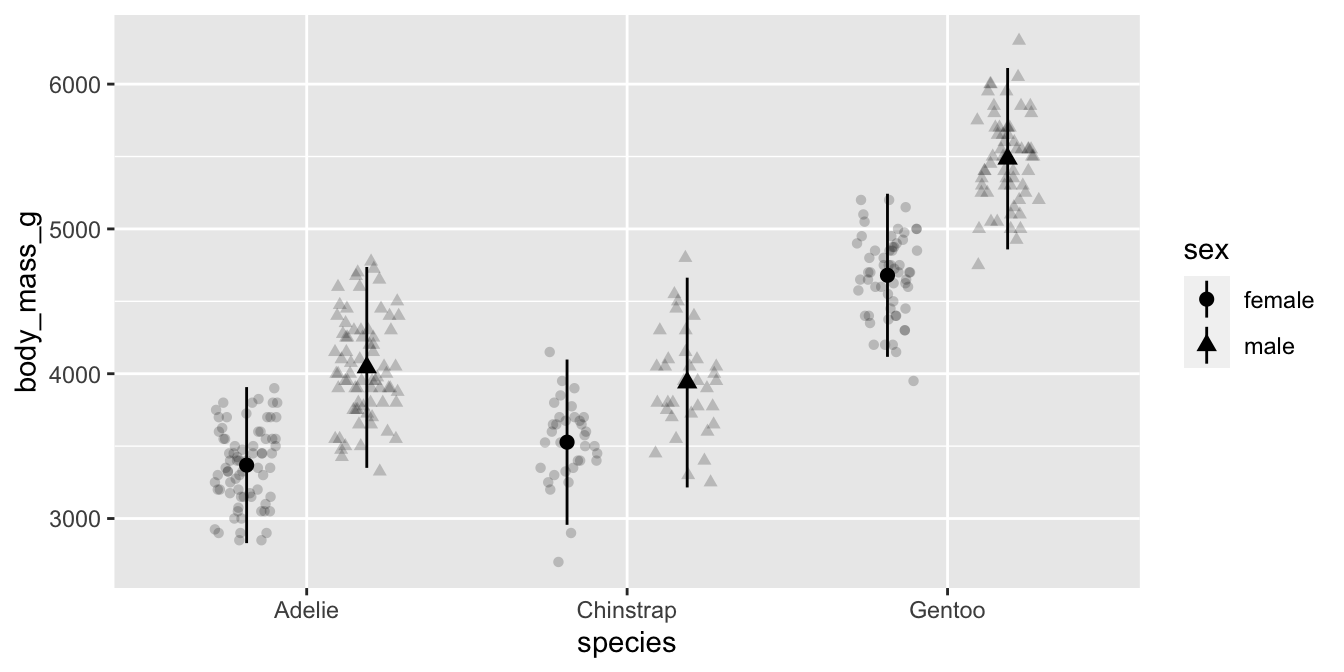
Applying what we learned

Axes
Axes
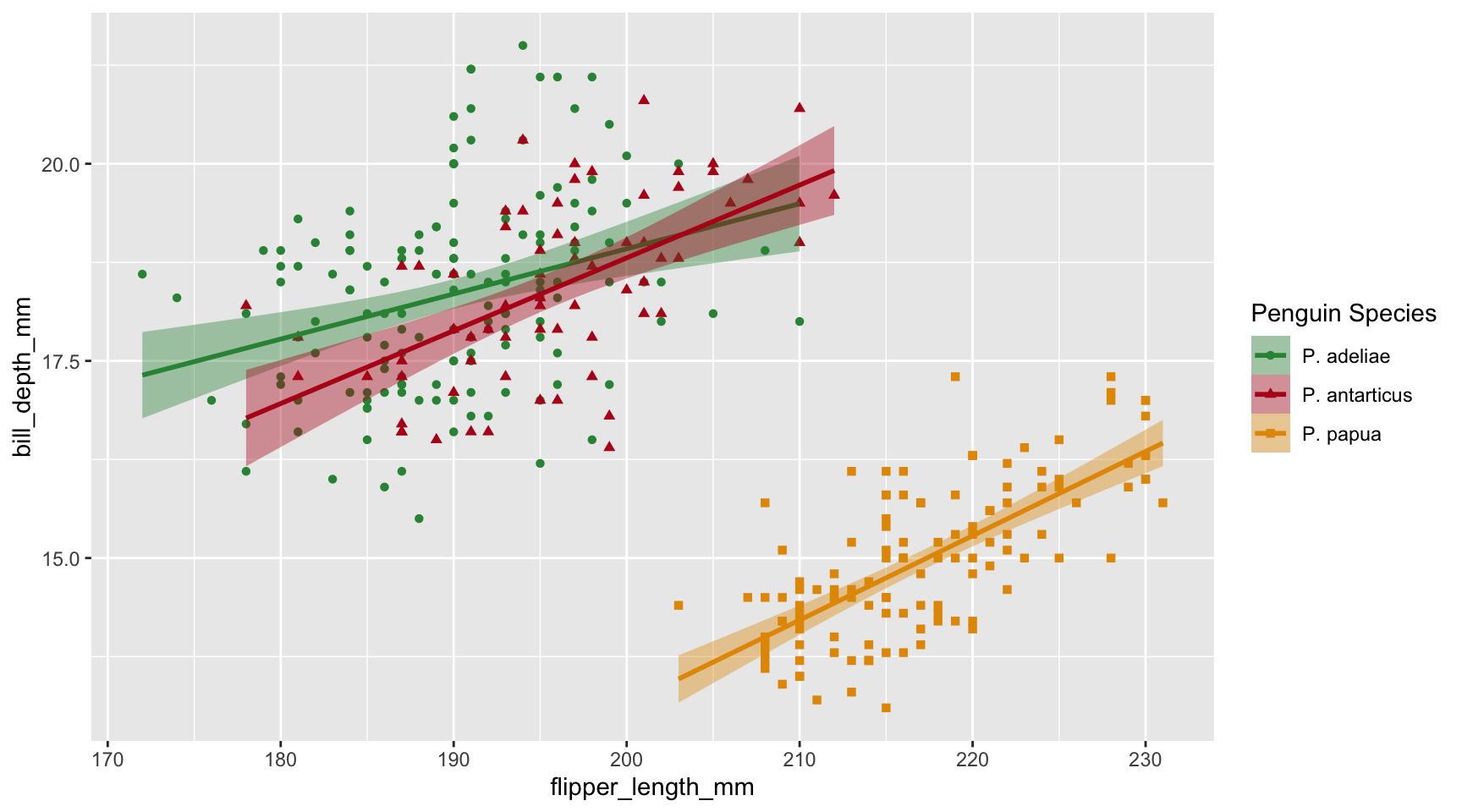
Axes are also a type of scale. In p1 the x-axis corresponds to scale_x_discrete() and the y-axis corresponds to scale_y_continuous().
Custom labels
Use what we learned before to customize the categorical x-axis labels in p1!

Custom labels
If you only want to change the axis title, you can also do that in labs()
Custom breaks
Change the (approximate) number of breaks with n.breaks=

Custom breaks
Specify breaks exactly with breaks=

Themes
Complete themes
There are several complete themes built-in to ggplot2, and many more available from other packages such as ggthemes.
Fonts
You can customize font size and family with complete themes.
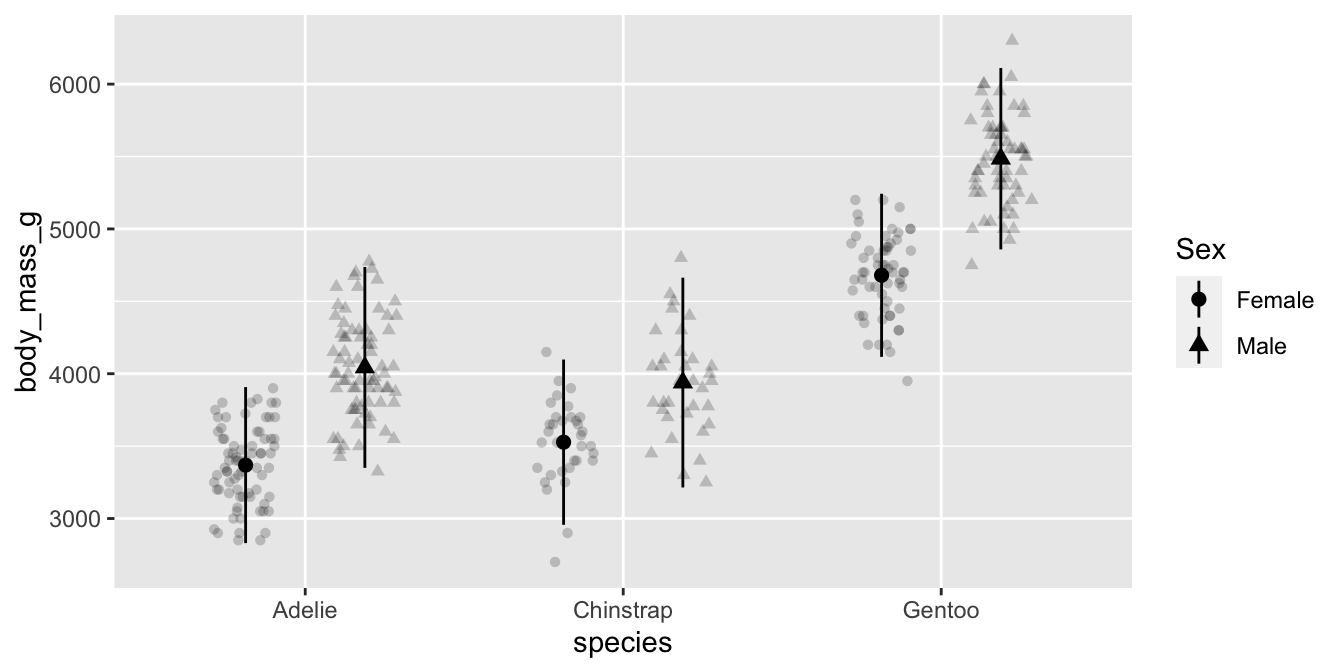
Custom themes
Customizing themes “manually” involves knowing the name of the theme element and it’s corresponding element_*() function.

Custom themes
It’s best to find a built-in theme_*() function that gets you most of the way there and then customize with theme()
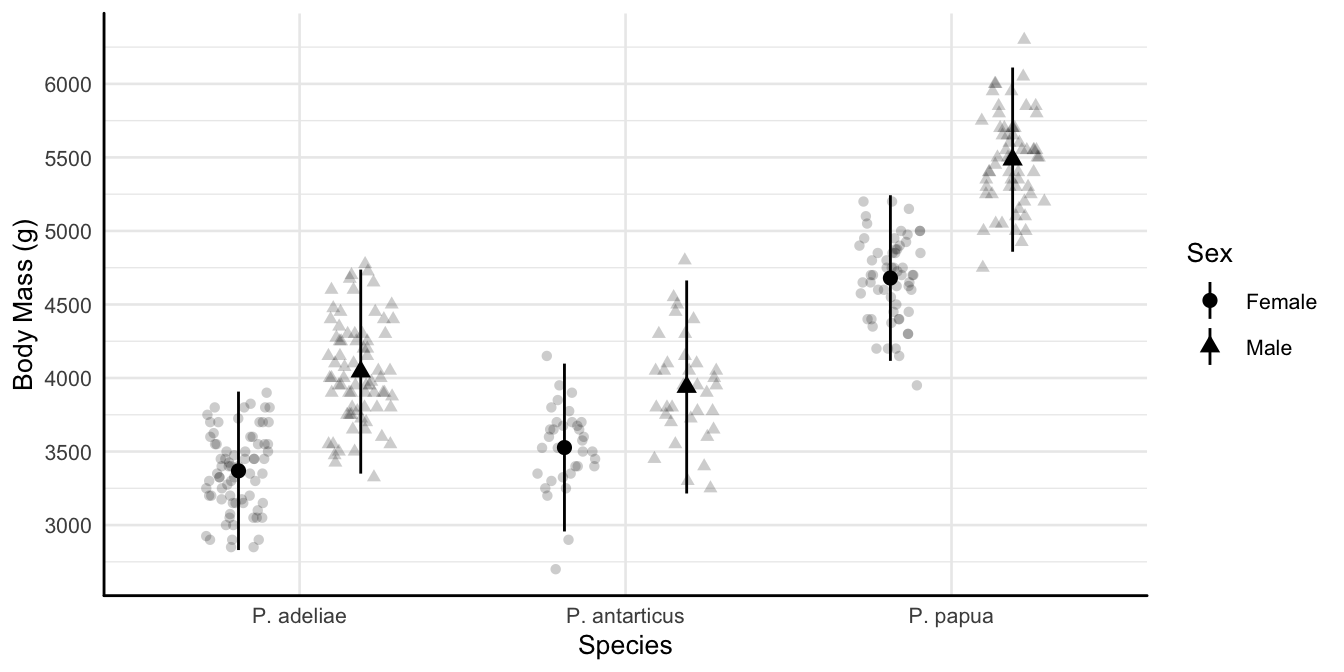
Custom themes
Activity
Name some things about the appearance of p1 that you want to change and we’ll figure it out together!
Tip
Check the examples in the help page for theme() https://ggplot2.tidyverse.org/reference/theme.html to figure out the names of theme elements
Re-using custom themes
You can save a custom theme as an R object and supply it to your plots.

Re-using custom themes
Or you can set your theme as the default at the top of your R script
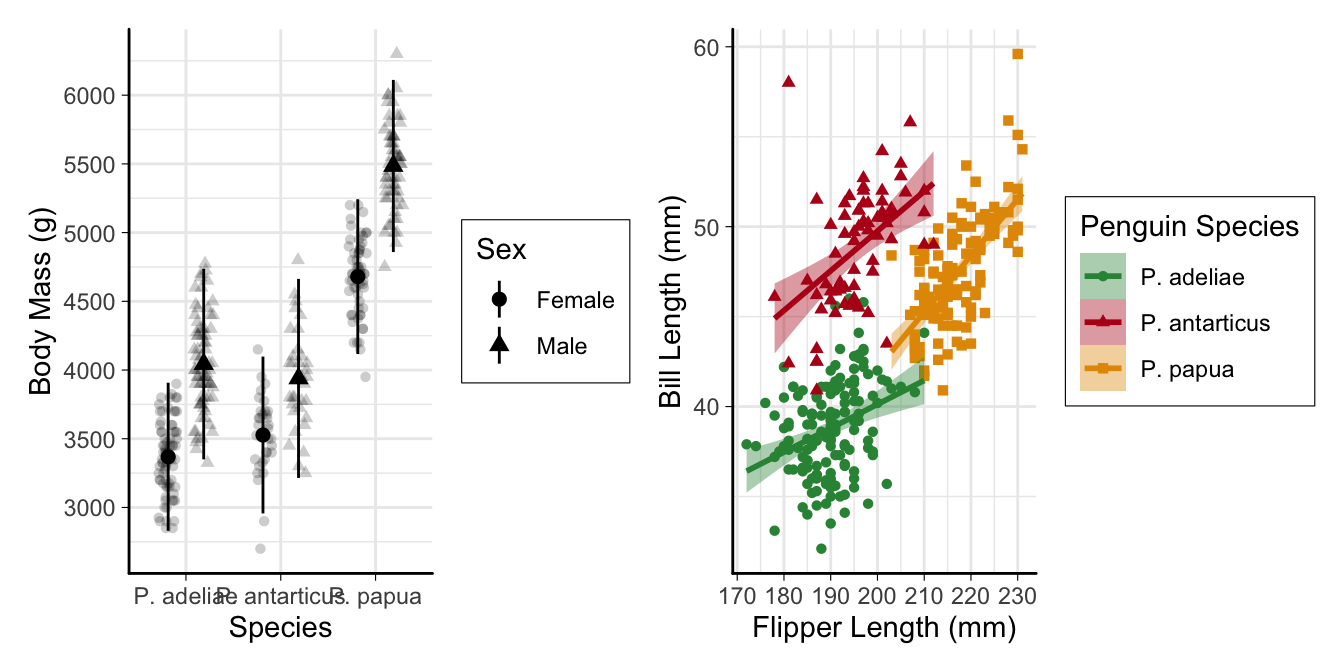
Multi-panel figures
Combine plots
The patchwork package makes it easy to combine ggplot2 plots
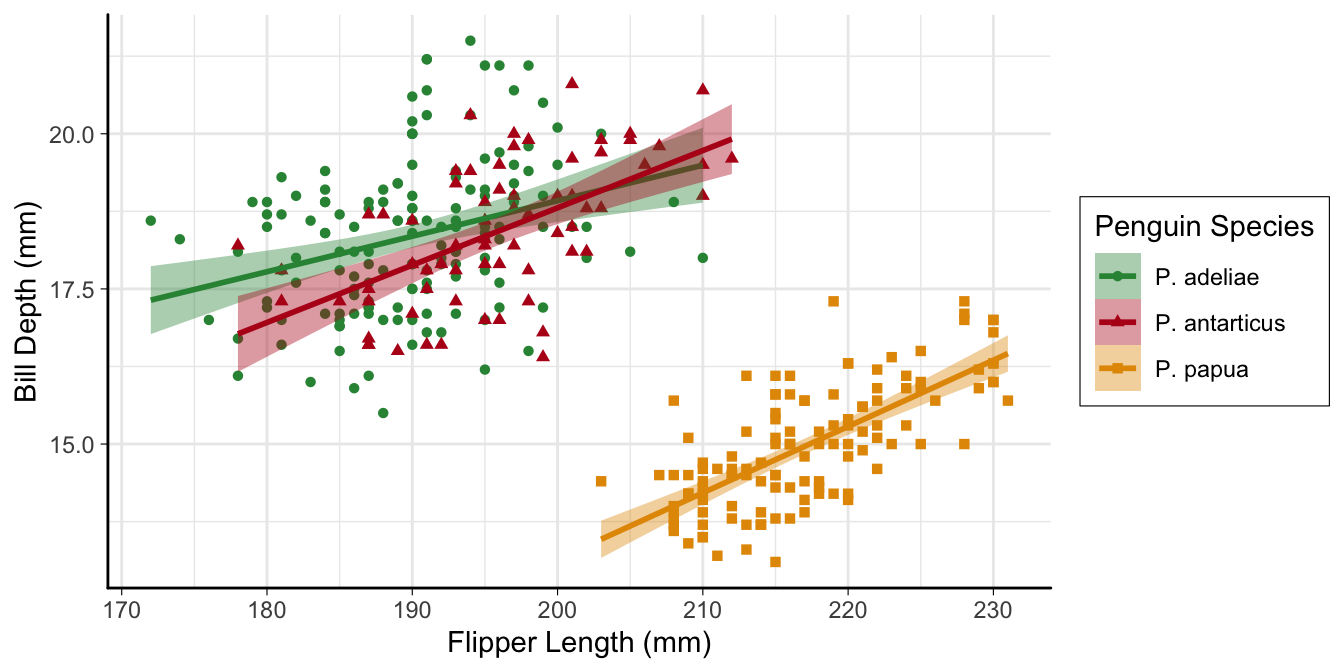
Control layout
Multi-panel figures
-
plot_layout(guides = "collect")combines duplicate legends -
plot_annotation(tag_levels = "a")adds labels to sub-plots
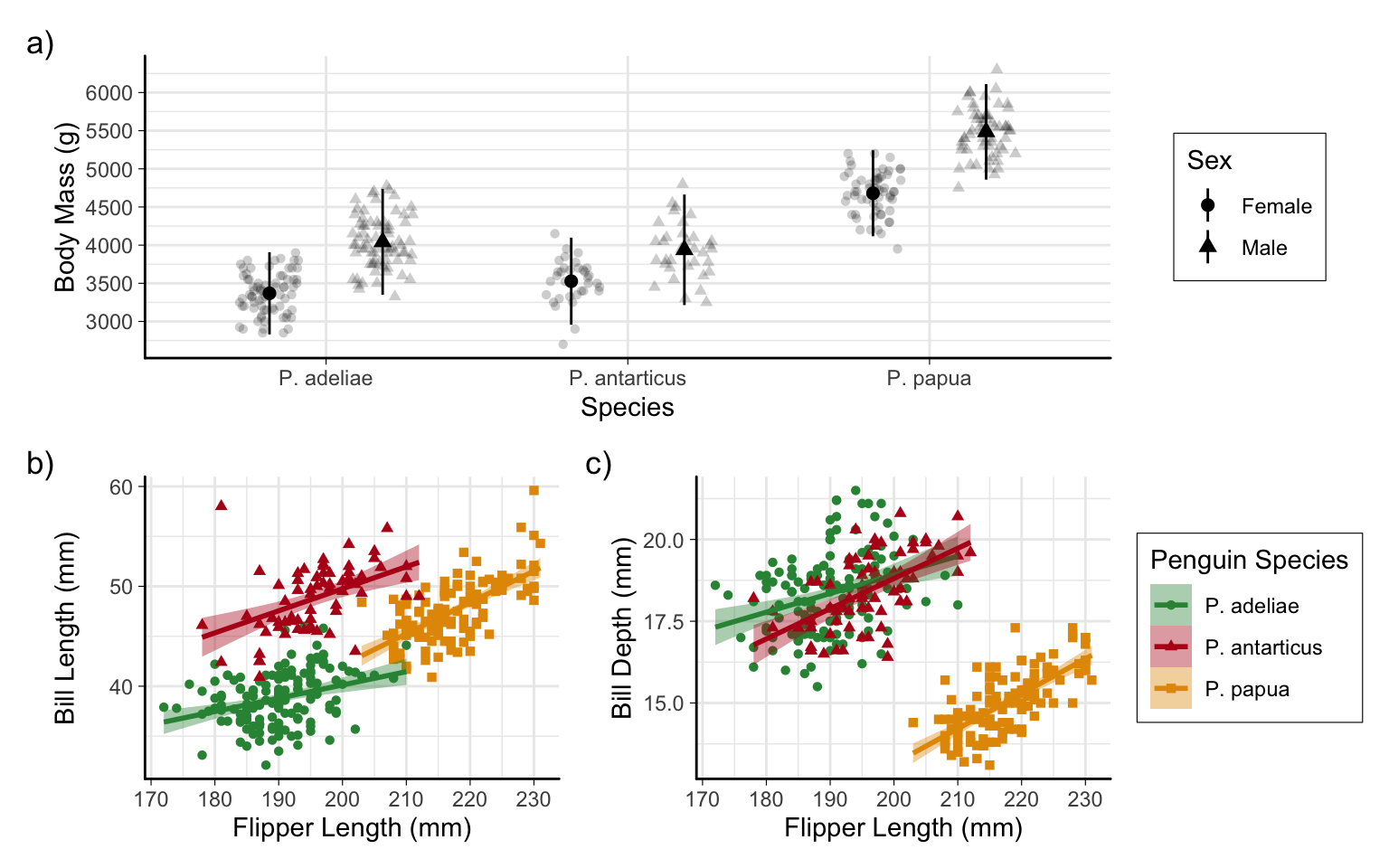
Saving plots 💾
Saving plots
If you know the dimensions, it’s good to save plots early on and adjust theme to fit.
Raster vs. Vector
- Raster images (e.g. .jpg, .png, .tiff) are made of pixels and the resolution can vary.
- Vector images (e.g. .svg, .eps) are not made of pixels and don’t have a resolution.
- Vector formats should be used whenever possible

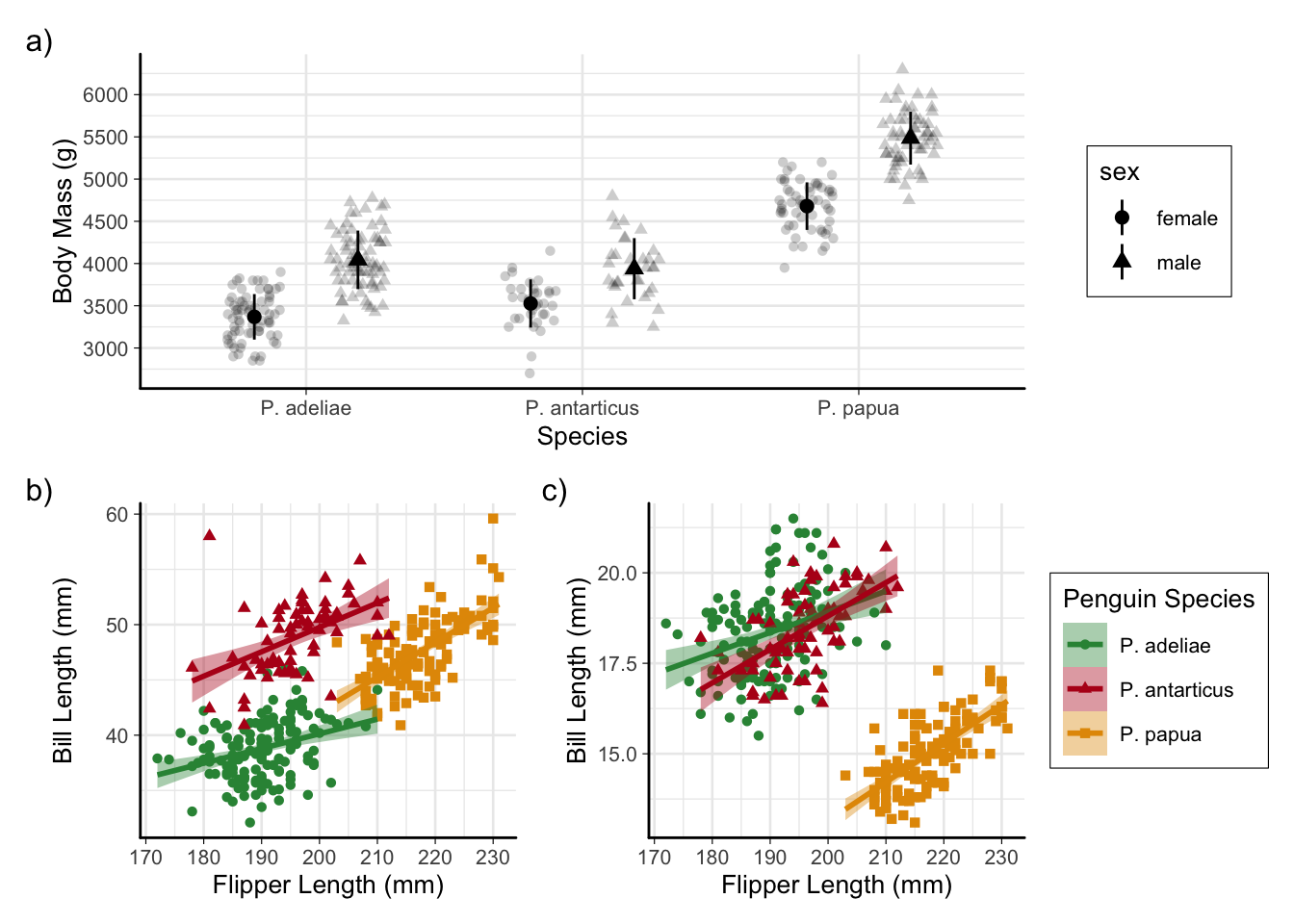
Finished product!
library(tidyverse) #includes ggplot2
library(palmerpenguins) #for data
library(patchwork) #multi-panel figures
# Custom theme
my_theme <-
theme_minimal(base_size = 10) +
theme(
axis.line = element_line(linewidth = 0.5, lineend = "round"),
axis.ticks = element_line(linewidth = 0.2),
legend.background = element_rect(linewidth = 0.2)
)
theme_set(my_theme)
# plot 1
## Custom function for stat_summary
mean_sd <- function(x) {
data.frame(y = mean(x), ymin = mean(x) - sd(x), ymax = mean(x) + sd(x))
}
## For labeling with latin names
scinames <- c("Adelie" = "P. adeliae",
"Chinstrap" = "P. antarticus",
"Gentoo" = "P. papua")
p1 <-
ggplot(penguins |> filter(!is.na(sex)),
aes(x = species, y = body_mass_g, shape = sex)) +
geom_point(alpha = 0.2,
position = position_jitterdodge(dodge.width = 0.75)) +
stat_summary(fun.data = mean_sd,
position = position_dodge(width = 0.75)) +
scale_x_discrete(labels = scinames) +
scale_y_continuous(n.breaks = 12) +
labs(x = "Species", y = "Body Mass (g)")
# Color palette for plot 2 and 3
my_cols <-
c("Chinstrap" = "#B60A1C", "Gentoo" = "#E39802", "Adelie" = "#309143")
p2 <-
ggplot(penguins,
aes(
x = flipper_length_mm,
y = bill_length_mm,
color = species,
fill = species,
shape = species
)) +
geom_point() +
geom_smooth(method = "lm") +
scale_color_manual(
values = my_cols,
labels = scinames,
aesthetics = c("color", "fill")
) +
scale_shape_discrete(labels = scinames) +
labs(
color = "Penguin Species",
shape = "Penguin Species",
fill = "Penguin Species",
x = "Flipper Length (mm)",
y = "Bill Length (mm)"
)
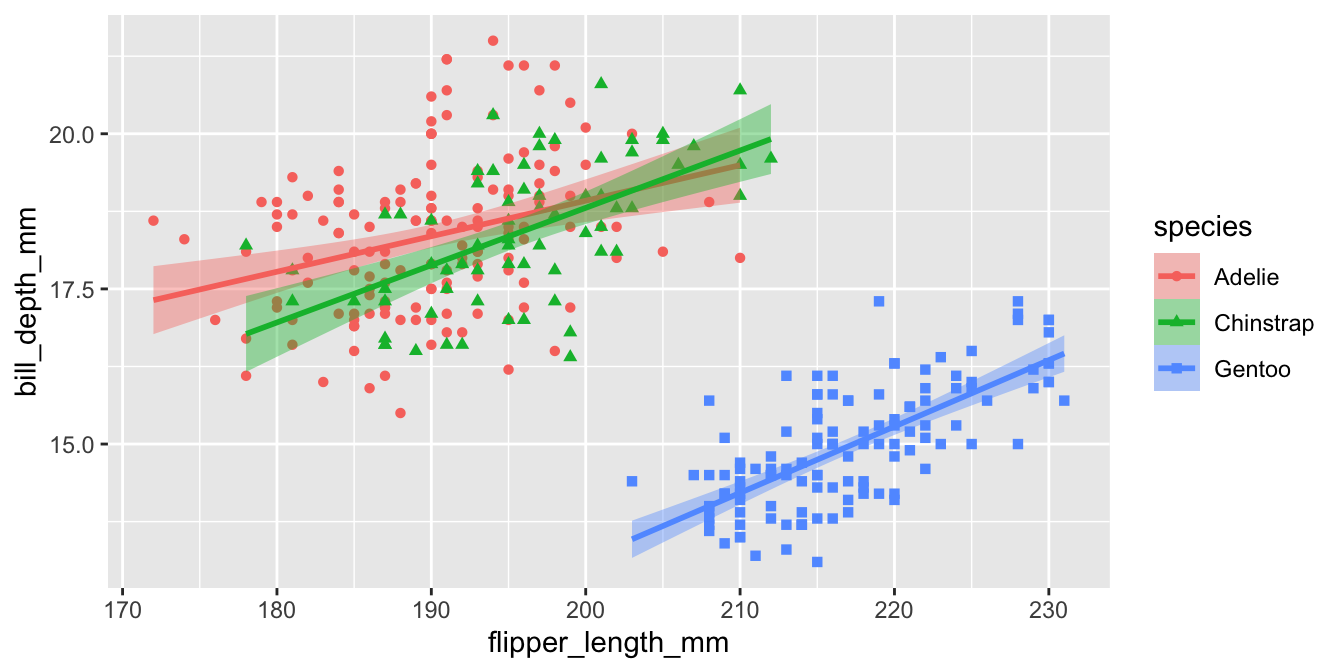
p3 <-
ggplot(penguins,
aes(
x = flipper_length_mm,
y = bill_depth_mm,
color = species,
fill = species,
shape = species
)) +
geom_point() +
geom_smooth(method = "lm") +
scale_color_manual(
values = my_cols,
labels = scinames,
aesthetics = c("color", "fill")
) +
scale_shape_discrete(labels = scinames) +
labs(
color = "Penguin Species",
shape = "Penguin Species",
fill = "Penguin Species",
x = "Flipper Length (mm)",
y = "Bill Length (mm)"
)
# combine into multi-panel figure
p_combined <-
p1 /
(p2 + p3 + plot_layout(guides = "collect")) +
plot_annotation(tag_levels = "a", tag_suffix = ")")
p_combinedFinished product!
Getting help
The
ggplot2documentation itself, in particular the examples on thetheme()page and the FAQ on customization.The R Graph Gallery has a wide variety of plots with R code walkthroughs.
The
ggplot2book: https://ggplot2-book.org/
You can always come by our drop-in hours to ask questions as well!
Part 3 in two weeks!
“Exploring the wide world of ggplot2 extensions”
🗓️ June 26
⌚️ 11:00am–1:00pm